
The second part of Bob Goldberg's Teaching Program, HC70AL - Gene Discovery Laboratory, provides students who have taken HC70A with an original, hands-on research project using state-of-the-art genomics technology. The Gene Discovery Laboratory teaches students who have never worked in a laboratory how to carry out an original research project, analyze data critically, and communicate results to their peers. Non-science students from majors as diverse as economics, communications, and classics, team up with beginning life science students in an intense ten-week effort to identify genes that control seed development. Bob Goldberg's goal is to give undergraduates who are headed to careers in the arts, business, law, and public service a unique insight into what scientists do, the laboratory life and culture, and experiencing how research is carried out. An additional objective is to introduce entering life science students to the excitement of discovery, providing them with a strong foundation for future undergraduate research efforts and beyond. HC70AL students work side-by-side with postdocs, graduate students, and technicians in Bob Goldberg's laboratory, using Arabidopsis as a model organism to identify genes required to "make a seed." Students are divided into research teams that are lead by peer laboratory mentors who are third- and fourth-year life science majors. HC70AL is organized into four parts: (1) an open laboratory that formally meets twice a week in which students use PCR, DNA sequencing, microarrays, RT-PCR, bioinformatics, and microscopy to identify and study knock-outs in transcription factor genes known to be active in seed development, (2) a weekly lab meeting and seminar (Fiat Lux MCDB19) in which students present their results and discuss ethical issues related to genetic engineering and research in general, (3) a weekly lab dinner that provides an informal setting for students to interact with Bob Goldberg and members of his research group, and (4) an all-class symposium and lab-coat ceremony at the end of quarter in which students formally present their results and are given a lab coat embroidered with their names. During the past seven years, 61 students participated in the Gene Discovery Laboratory, and knock-outs in over 103 Arabidopsis transcription factor genes have been identified and studied.
GENE DISCOVERY LABORATORY
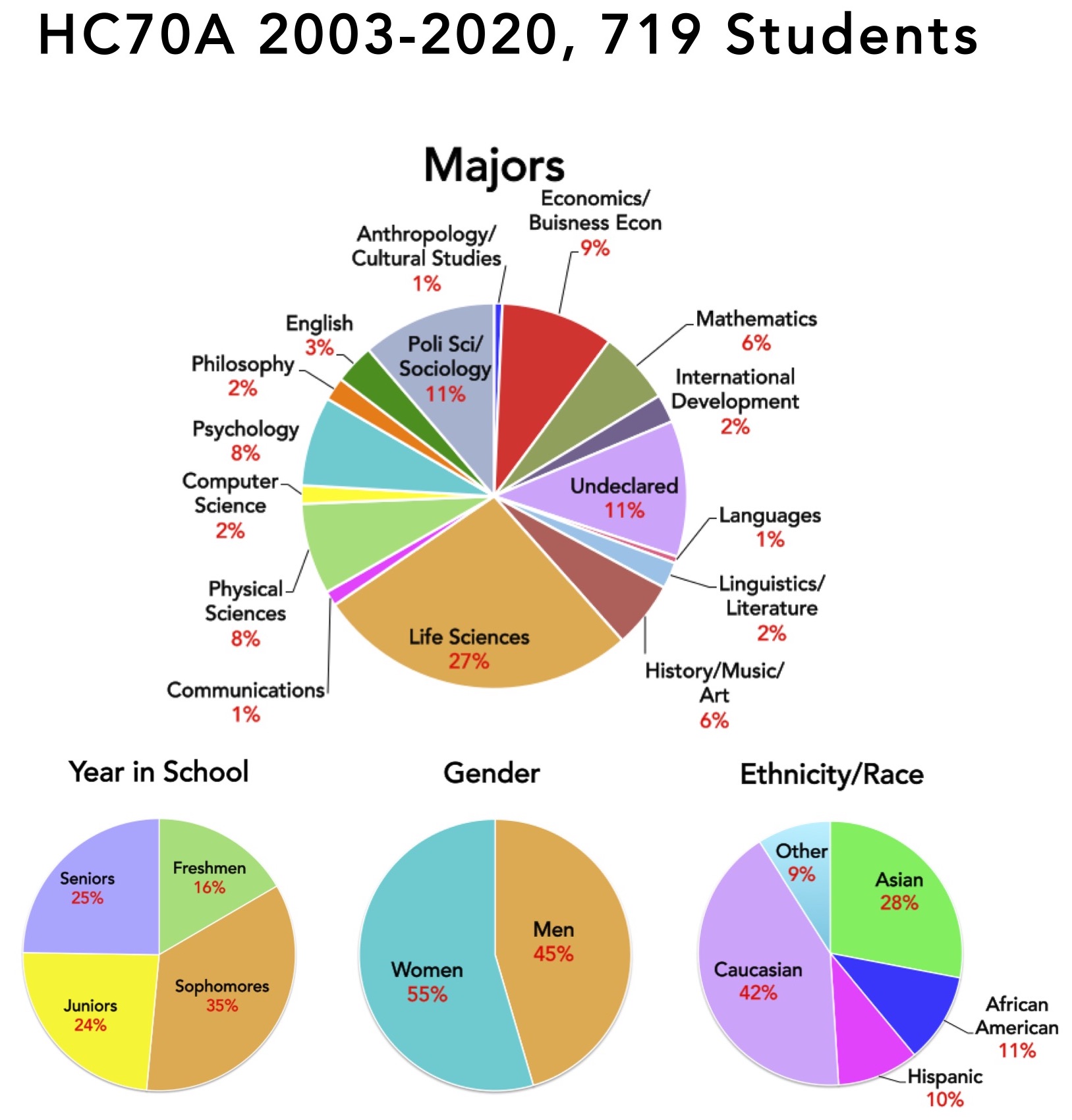
Distribution of Students That Have Taken the HC70AL Gene Discovery Laboratory
One of the unique aspects of HC70AL is the breadth of undergraduate majors and levels represented by students in the class. Freshman work side-by-side with seniors, and life science students that will become molecular biology, microbiology, and biochemistry majors team up with philosophy, business, and communication majors, among others, in carrying out their experiments. Kyoto University students also participate in HC70AL, providing an novel cross-cultural laboratory experience for everyone involved. During the past six years, 61 students took HC70AL, representing 15 different majors. Approximately 30% of the students were undeclared, and approximately 45% of the students were non-science students, and the remainder entering life science students who had declared a major but were just beginning the core curriculum required of all life science students at UCLA. The pie charts shown on this page summarize the HC70AL student population over the past six years.
GENE DISCOVERY LABORATORY QUESTIONS, APPROACHES, AND RESULTS
Research carried out in Bob Goldberg's laboratory has identified transcription factor (TF) genes that are active at specific stages of seed
development using genomics with a large number of microarray experiments. These studies profiled gene activity in whole seeds, as well as in
every tissue, compartment, and region of both Arabidopsis and soybean seeds throughout development using laser capture microdissection (LCM)
technology. A description of this NSF-funded research project and results of all the microarray experiments are available on the project website
(click here to visit the project website). HC70AL undergraduates have used reverse genetics to identify and characterize knock-outs in seed-stage-
specific and/or seed-tissue-specific TF genes identified in the microarray experiments, and begin to answer the question of what role these TF
genes perform in seed development. The Arabidopsis knock-out lines were generated at the Salk Institute (click here to visit Salk website). In
addition, HC70AL students cloned and sequenced the promoters of each TF gene they investigated to give them hands-on experience in genetic engineering
- the focus of their HC70A experience - and to use in promoter-reporter gene fusion experiments in the future to study the location of TF gene
transcription within the seed. During the past six years, 103 Arabidopsis seed TF gene knock-out lines have been identified and characterized by
HC70AL students, who had never held a pipette before.
In 2009, HC70AL students carried out a group project that sequenced the Scarlet Runner Bean (SRB) genome using high-throughput sequencing technology.
1.1Gb of SRB DNA sequences was obtained and assembled into 278,000 contigs. HC70AL students annotated 242Kb DNA sequences and identified 36 genes
that were supported by EST sequences in our SRB EST database.
UNDERGRADUATE RESEARCH TOOLS DEVELOPED FOR THE GENE DISCOVERY LABORATORY
Gene Discovery Laboratory Lab Manual
A comprehensive set of experiments and protocols was written for undergraduate laboratories using genomics to study the function and activity of plant genes. This lab manual was designed to be used by both non-science and science majors. The Gene Discovery Lab Manual introduces students to basic molecular biology techniques and contains week-by-week experiments that allow undergraduates to use reverse genetics to identify T-DNA knock outs in Arabidopsis genes, describe the expression pattern of wild-type and mutant Arabidopsis genes, amplify and clone Arabidopsis gene promoters, and use microscopy to study wild-type and mutant seeds phenotypes. Detailed protocols are provided for DNA sequencing, PCR, RT-PCR, bioinformatics to characterize gene structure and T-DNA insert sites, planting seeds and growing Arabidopsis plants, isolating DNA and RNA, and using Nomarski optics to study seed and embryo phenotypes, among others.
Interactive Lab Webbook
An web-based interactive laboratory webbook was created that allows students and instructors real-time viewing of all experiments and results obtained in the Gene Discovery Laboratory. Students use the webbook to organize, summarize, and post their research results. In addition, protocols can be uploaded to the webbook and linked to each experiment. The webbook uses a relational data base and is designed to be used in any undergraduate laboratory course or research laboratory.
GENE DISCOVERY LABORATORY FINAL SYMPOSIUM AND LAB COAT CEREMONY
At the end of the quarter, undergraduates give PowerPoint presentations of their research results at the final class symposium. Groups of students present an overview of seed development and the features of each transcription factor family investigated during the quarter, providing an introduction and background information for the symposium. Each student then gives a ten minute talk on their specific research results. At the end of the symposium Bob Goldberg presents a cake to the class decorated with a relevant design, and presents each student with a lab coat embroidered with their name, class year, and Goldberg Laboratory in honor of their hard work and participation in the gene Discovery Laboratory.
WHY STUDY SEEDS?
Seeds are one of the most important sources of food for human and animal consumption. Over half of the calories consumed by humans world wide is obtained from either
seeds or seed by-products (e.g., rice, wheat, maize, soybean). In addition, most major crop plants are generated directly from seeds.
Seeds contain a complex collection of cells, tissues, and organs with distinct origins and developmental fates that ensure the survival of higher plants. The major
events that occur during seed development are shown schematically in the figures on this web page. Seed development begins with a unique double fertilization process.
One sperm cell unites with the diploid central cell of the embryo sac, giving rise to a triploid endosperm tissue that nourishes the embryo during the early stages of
seed development. The other sperm cell unites with the egg cell giving rise to the zygote that initiates embryo formation. At maturity, a seed contains a fully
differentiated embryo that contains one or two cotyledons and an axis. The cotyledons accumulate storage reserves that are used after germination by the developing
seedling. The axis contains meristematic tissues that will give rise to the stem, roots, and leaves of the plant following germination. By contrast, the endosperm is
either fully absorbed by the embryo during seed development (e.g., Arabidopsis) or is consumed during germination and supports growth of the seedling until photosynthesis
begins (e.g., maize). Proteins, oils, and starches that are stored in the endosperm and cotyledons are the major sources of food provided by seeds for human and animal consumption.
Little is known about the processes that regulate the complex events that are required to coordinate both endosperm and embryo formation during seed development. That is,
the genes and regulatory networks that control seed development are not well understood. Because seeds are an important source of human nutrition, Bob Goldberg's laboratory
hopes to identify all of the genes required to "make a seed." He wants to use this information to engineer crops to produce more seeds and/or bigger seeds and, ultimately,
increase the human food supply. Undergraduates who participate in the HC70AL Gene Discovery are helping to facilitate this goal by using functional genomics to identify
genes that play a major role in controlling seed development.
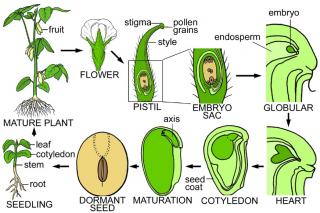
A schematic diagram of the soybean plant's life cycle